[Hyperendemie und Genotypenvielfalt des Hepatitis-B-Virus bei Patienten des Lehrkrankenhauses der Universität Abuja, Nigeria]
Chinwe Ndidi Ugwu 1Amos Dangana 2
Hassan Suleiman Chunta 3
Ishaku Akyala Adamu 3
Nanpon Miri 2
Mangpin Leviticus Dansura 2
Bwede Eugene Samuel 2
Felix Villeng Gagari 2
Johnson Adeyemi Ojo 2
Nyiri Miriam Gyang 4
Nkiruka Lynda Uzoebo 2
Helen Daniel Nanbol 5
Gabriel Bernice Nyinishu 6
Aisha Daminso Barde 4
Idris Nasir Abdullahi 7
1 Biorepository Laboratory, Nigeria Centre for Disease Control and Prevention, Abuja, Nigeria
2 National Reference Laboratory, Nigeria Center for Disease Control and Prevention, Abuja, Nigeria
3 Global Health and Infectious Diseases Control Institute, Nasarawa State University, Keffi, Nigeria
4 Mega Laboratory, Nigeria Center for Disease Control and Prevention, Abuja, Nigeria
5 School of Medical Laboratory Science, Plateau State College of Health Technology, Pankshin, Nigeria
6 Sherrif Integrated Healthcare Limited, Abuja, Nigeria
7 Department of Medical Laboratory Science, Faculty of Allied Health Sciences, College of Medical Sciences, Ahmadu Bello University, Zaria, Nigeria
Zusammenfassung
Einleitung: Die Infektion mit dem Hepatitis-B-Virus (HBV) ist nach wie vor ein weltweites Problem der öffentlichen Gesundheit, insbesondere in den afrikanischen Ländern südlich der Sahara, da die medizinische Versorgung und die Erforschung der genetischen Epidemiologie unzureichend sind. Ziel dieser Studie ist es, die Häufigkeit von HBV-Antigenen, -Antikörpern und -Genotypen bei fiebrigen Patienten zu bestimmen, die das Lehrkrankenhaus der Universität Abuja (UATH), Nigeria, besuchen.
Methode: An der Querschnittsstudie nahmen 100 Patienten teil, deren Blutproben entnommen und mittels Lateral Flow Assay auf HBV-Oberflächenantigen (HBsAg) und vier weitere strukturelle Antigene und Antikörper untersucht wurden. Alle HBsAg-positiven Proben wurden mittels typspezifischer PCR genotypisiert. Mit strukturierten Fragebögen wurden die soziodemografischen Variablen der Patienten erfasst.
Ergebnisse: Die HBsAg-Seropositivität betrug 31%. Die Verteilung der HBV-Genotypen war wie folgt: Der Genotyp E war vorherrschend (22,6%), gefolgt vom Genotyp B (16,1%). Von den HBsAg-positiven Personen waren alle HBsAb-seronegativ, 3,2%, 74,2% und 90,3% waren HBeAg-, HBeAb- bzw. HBcAb-seropositiv. Die Genotypen B, C und D wurden bei 16,1%, 3,2% bzw. 3,2% nachgewiesen. Gemessen an der Anzahl der HBV-Genotypen pro Person hatten 9,7% einen Genotyp, 16,1% hatten zwei Genotypen und 74,2% hatten drei Genotypen. Der Bildungsabschluss war signifikant mit der Anzahl der dreifachen HBV-Genotypen pro Person verbunden (p=0,04).
Schlussfolgerung: Es wurde eine sehr hohe Seroprävalenz von HBV festgestellt, wobei der Genotyp E vorherrschte. Das Vorhandensein mehrerer HBV-Genotypen innerhalb eines Wirts wurde zum ersten Mal in Nigeria festgestellt. Das weist auf die genetische Heterogenität von HBV in Nordnigeria hin mit möglichen Auswirkungen auf die verfügbaren Kontrollmaßnahmen.
Schlüsselwörter
HBsAg, HBV genotype, endemisch, Nigeria
Introduction
Hepatitis B virus (HBV) infection is a major global health concern, causing significantly higher morbidity and mortality in developing countries [1]. In Nigeria, HBV is reported to be the most common cause of liver disease [2]. Individuals who test HBsAg-seropositive need to have other hepatitis B markers (anti-HBs, HBeAg, anti-HBe, and HBV DNA) evaluated to determine the stage and severity of infection [3].
HBV is endemic in Nigeria [4]. However, there is limited data on the comprehensive evaluation of hepatitis B markers to provide critical data to optimize and assess the impact of current prevention and control strategies, including disease surveillance and diagnoses, vaccination policies, and management [1]. Nigeria has a very high risk of HBV infection because of the country’s poor immunization rates and the fact that up to 75% of the population will be exposed to the virus at some point in their life [5].
Before the characterization of HBV genotypes, strains were distinguished by serological analysis, based on the immunoreactivity of an antibody to a limited number of amino acids in the major surface antigen, HBsAg [6]. Subsequently, HBV now exists in 10 known genotypes (A–J) and 40 subgenotypes based on an intergroup divergence of 8% or 4%, using the gene-S sequence [7]. The natural course of an HBV infection may be influenced by the genotypes or subgenotypes of the virus, but the identification of a particular genotype or subgenotype is not necessary for the administration of antiviral medication [8]. However, given that the genotypes are known to differ across geographic regions and have a high correlation with ethnicity, they have been proposed as valuable epidemiological identifiers [9].
Acute self-limiting hepatitis is linked to genotype D, chronic active hepatitis is linked to genotype A, and liver cancer is linked to genotype B [8], [9]. Despite being widespread, genotype A is mostly found in Northern Europe, Central Africa, and North America [7], [10]. Southeast Asia and the Far East have reported cases of HBV infections due to genotypes B and C [11]. Genotype D is mostly found in the Mediterranean area, but is distributed throughout the world [11].
West Africans and Native Americans primarily have genotypes E and F, respectively. Lately, genotype H has been discovered in Central America, and genotype G has been discovered in the United States and France [12]. Genotypes I and J are understudied. However, Asian genotypes I and J are thought to be the product of recombination events with other genotypes [12].
If HBsAg persists for more than 6 months, spontaneous clearance is very improbable, and the infected individual is a chronic HBV carrier [13]. However, during the acute phase of infection, following HBsAg detection, other viral markers can be easily detected, including DNA polymerase and HBeAg [13]. HBeAg appears shortly after the appearance of HBsAg and disappears within several weeks as acute hepatitis resolves [14]. Its presence in the serum correlates with the presence of viral replication in the liver and HBsAg detection, while its disappearance, associated with anti-HBe detection, is considered a sign of the absence of viral replication and spontaneous resolution of acute infection [14]. Anti-HBc IgM antibodies are detectable at the outset of clinical disease; as the infection evolves, IgM anti-HBc levels gradually decline, often becoming undetectable within 6 months, and IgG class predominates, remaining for a long period (sometimes life-long) at detectable levels [15]. IgG anti-HBc is correlated with prior infection, whereas IgM anti-HBc often suggests recent or ongoing HBV replication [16]. In cognizance of the enormous clinic-epidemiological value of characterizing the serological markers and genotypes in HBV-infected individuals, this study aims to determine the frequency of HBV antigens, antibodies, and genotypes among febrile patients attending the University of Abuja Teaching Hospital, Nigeria.
Materials and methods
Study center
This cross-sectional study was performed at the University of Abuja Teaching Hospital (UATH) Gwagwalada Municipal Area in the Federal Capital Territory (Abuja), Nigeria.
Study size and participants
A minimum sample size of 89 was calculated from the 6% seroprevalence of HBsAg previously reported in the same study area [17]. All patients who consented to participate were randomly enrolled at the various departments and clinics of the University of Abuja Teaching Hospital. Pregnant women, diabetic patients, and people living with HIV and AIDS at the time of sample collection were excluded. Parents provided informed consent on behalf of the children.
Ethics approval
The study was approved (No.: FCT/UATH/HREC/14723) by the Human Research Ethics Committee (HREC) of the University of Abuja Teaching Hospital Gwagwalada (UATH), Abuja, Nigeria. All authors gave informed consent before enrolment into the study. Sample collection and data collation from the study participants were according to the Declaration of Helsinki.
Sample collection and processing
Each participant had a total of five milliliters (5 mL) of whole blood drawn and separated into 3 mL in an EDTA tube and 2 mL in a plain tube. To extract gDNA, 2 ml (microliters: 200 µL) of plasma were extracted directly from the EDTA-anticoagulated blood. The EDTA-anticoagulated blood was centrifuged at 5,000 revolution per minute for five minutes to remove the plasma and separate it from the red blood cells. After that, the transparent plasma layer was placed in cryovials and kept cold until analysis. Furthermore, the sera were extracted from the clotted whole blood in the plain tube. The sera were used for HBV serological tests, while the plasma samples were used for HBV DNA testing by PCR at the National Reference Laboratory (NRL), Nigeria Center for Disease and Prevention.
Laboratory analytical procedures
Detection of HBV serologic markers (HBsAg, HBsAb, HBeAg, HBeAb and HBcAb) was performed using the HBV-5 panel test kit (Abbott Laboratories Biotech Co. Ltd) was used. The test was based on (a) the direct antigen-antibody-antigen “Sandwich” method for HBsAg, HBsAb, HBcAg and HbeAg, and (b) the indirect antigen-antibody-antibody assay for HBcAb. The sandwich immunochromatographic strips method contains gold nanoparticles pre-coated with monoclonal antibodies specific to HBsAg, HBeAg, HBc IgM, or antigen specific to HBs recombinant monoclonal antibodies detectable in serum samples. The test was conducted and the result interpretedout according to the manufacturer’s instructions.
The HBV DNA was extracted from the plasma of HBsAg-positive patients using a Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s recommendations. The HBV genotypes were detected using the nested PCR using the primers (Table 1 [Tab. 1]).
Table 1: Primers for HBV genotyping (adopted from Naito et al. [34])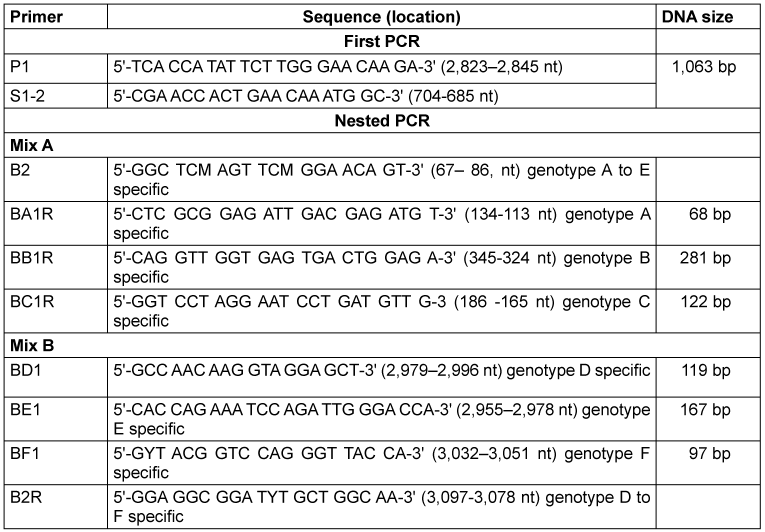
Nested Multiplex PCR was used for HBV DNA amplification using the universal primers (P1 and S1–2) for the outer primers and two different mixtures containing type-specific inner primers. The first PCR was carried out in a final reaction volume of 25 µl containing 2.5 µl of 1XPCR buffer, 1.0 µl of MgCl2, 0.8µl of forward primer, 0.8 µl of reverse primer, 0.8 µl of dNTPs, 0.2 µl of Taq, 13.0 µl of PCR water, and 5.0 µl of DNA. The thermocycler (Bonn-Bad Godesberg, Germany) was programmed to first incubate the samples for 10 min at 95°C, followed by 40 cycles of 94°C for 20s, 55°C for 20s, and 72°C for 1 min. Two second-round PCRs were performed for each sample, with the common universal sense primer (B2) and mix A for types A through C, and the common universal antisense primer (B2R) and mix B for types D through F. A 1-µl aliquot of the first PCR product was added into two tubes containing the second sets of each of the inner primer pairs, each of the deoxynucleotides, Taq and PCR buffer, as in the first reaction. These were amplified for 40 cycles, incubating at 95°C for 10 min, 20 cycles of amplification at 94°C for 20 s, 58°C for 20 s, 72°C for 30 s, an additional 20 cycles of 94°C for 20 s, 60°C for 20 s, and 72°C for 30 s. Genotype-specific DNA bands were used to identify the HBV genotypes.
The DNA from the PCR products was separated on a 10% agarose gel, stained with SYBR green, and visualized under ultraviolet light.
Results
Out of the 100 patients tested, 31% were HBsAg-positive. Among the 31 HBsAg-positive individuals, most of them were ≤30 years of age (51.6%), 51.6% were females, and 48.4% were married (Table 2 [Tab. 2]). Regarding educational qualifications, a majority had tertiary education (67.7%). Most of the participants (90.3%) reported no alcohol consumption. Regarding medical history, 71.0% had no prior hepatitis B history. Notably, none of the participants (100%) had been vaccinated against HBV. Various forms of chronic comorbidity were reported in 48.4% of the participants (Table 2 [Tab. 2]).
Table 2: Demographic and risk factors in HBsAg-positive participants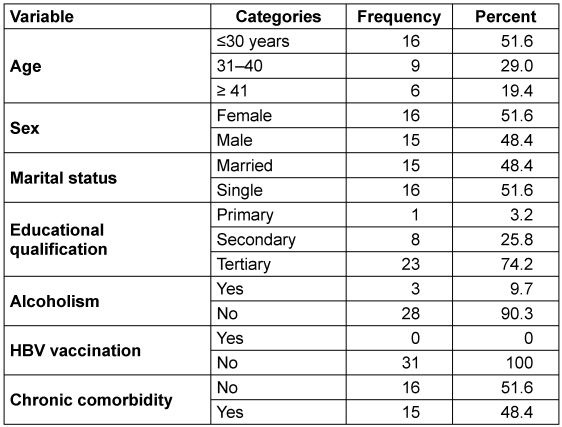
All 31 HBsAg-positive individuals tested negative for HBsAb (100%). For HBeAg, 3.2% tested positive. Moreover, 74.2% tested positive for HBeAb, and 90.3% tested positive for HBcAb (Table 3 [Tab. 3]).
Table 3: Hepatitis B serological markers in the HBsAg positive individuals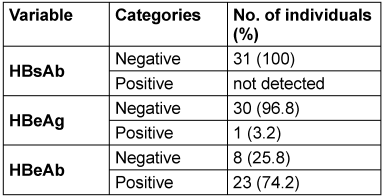
Of the 31 HBV-infected participants, HBV genotype E was predominant (22.6%). Genotypes B, C, and D were detected in 16.1%, 3.2%, and 3.2%, respectively. However, genotypes A and F were not detected at all (100.0%) (Table 4 [Tab. 4]). Concerning the number of HBV genotypes in an individual, 9.7% had a single genotype, 16.1% had double genotypes, and 74.2%, had triple genotypes.
Table 4: Frequency of Hepatitis B virus genotypes among HbsAg-positive individuals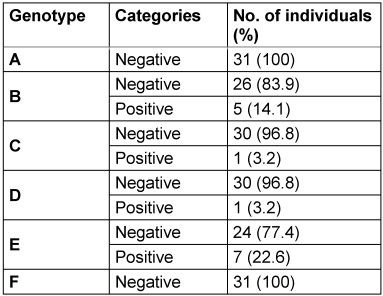
All participants within the age group ≥41 years had the highest percentage of triple HBV genotypes, followed by those ≤30 years (75%) (p=0.16). Both married and single individuals exhibit similar patterns in the number of HBV genotypes (p=0.92). Males had relatively higher (80%) triple HV genotypes than did females (68.8%) (p=0.47).
Educational qualification was significantly associated with multiple HBV genotypes per individual (p=0.04). In this regard, individuals with a secondary education had the highest frequency of triple HBV genotypes (48.4%) (Table 5 [Tab. 5]).
Table 5: Association between sociodemographic characteristics and triple HBV genotypes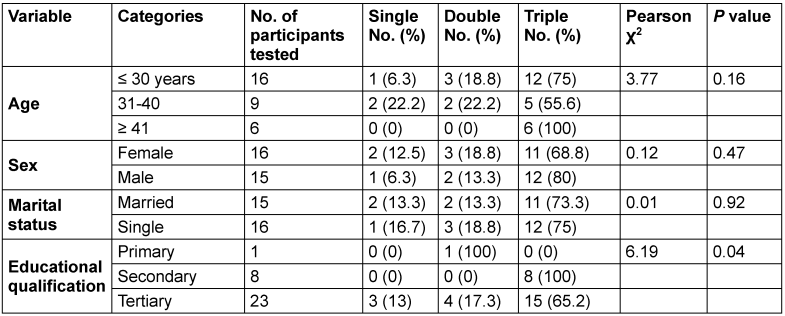
Discussion
In any given population, an HBsAg prevalence estimate of more than 8% is deemed high [18]. The high HBsAg rate in this study indicates that Nigeria is HBV hyperendemic [19]. Globally, the prevalence of chronic HBV infection varies, from less than 1% in low-endemicity areas to more than 30% in highly endemic areas, depending on sociodemographic factors, lifestyle, clinical conditions, and vaccination coverage [20]. Similar high HBV seroprevalence in Nigeria has previously been reported in 44.7% of healthy pupils, 32% of patients, 29.7% in chronically infected persons, and 51.9% in people living with HIV/AIDS [21], [22], [23], [24], [25]. It is important to remark that most cross-sectional studies reported HBsAg frequency ranges of 4–22% in Nigeria [1].
In a global meta-analysis of the prevalence of HBV, it was reported that most developing countries in sub-Saharan Africa and Southeast Asia are hyperendemic for HBV infection with range of 10-30% [26].
All participants in the present study were negative for HBsAb, indicating a lack of immunity from prior infection or vaccination. Hence, they all had active hepatitis B. Of these, only one individual was HBeAg, a marker of active replication in the liver cells. This implies that the individual could be quite contagious due to their potentially high viral loads [27]. HBeAb, an indicator of resolved infection or low replication, was identified in most HBsAg-seropositive individuals.
The predominance of HBV genotype E in the present study conforms with most previous reports in Nigeria in different subpopulations with varying clinical conditions [21], [28], [29], [30]. This indicates the establishment of HBV genotype E in Nigeria. Specifically, genotype E is prevalent in West and Central Africa [11].
When many HBV genotypes are present in the same host, it is referred to as an HBV mixture. This can happen as a result of super- or co-infection [31]. In the present study, many within-host dual or triple HBV genotypes were identified. These are due to clusters associated with multiple introductions into the studied populations. When two or more different HBV genotypes or sub-genotypes infect the same host cell, they exchange genetic material during replication, a process known as recombination [32]. This phenomenon could have an impact on clinical outcomes, such as the development of liver fibrosis, cirrhosis, and hepatocellular cancer [31]. To our knowledge, this is the first report of within-host multiple HBV genotypes in Nigeria. Similar findings were recently reported in individual pregnant women in Ghana [33]. The absence of genotypes A and F is consistent with their rarity outside specific geographic regions.
Very high seroprevalence of HBV was found and genotype E predominated. Multiple HBV genotypes per individual were identified for the first time in northern Nigeria. This underscores the genetic heterogeneity of the virus in this region and suggests potential implications for disease progression, treatment response, and vaccine efficacy. It is recommended to perform phylogenomic analyses of the predominant HBV genotypes E and B identified in the study center to understand the evolutionary trajectory within and outside the region. Although the sample size of this study was statistically adequate, larger cohorts are required to improve epidemiological relevance.
In conclusion, there is a need to maintain HBV vaccination programs in infancy and revaccinate individuals at high risk of infection in adulthood. Moreover, other preventive measures, such as safe sex practices and avoidance of sharing syringe needles and sharp objects, should be encouraged.
Notes
Ethical approval
This study was approved (No.: FCT/UATH/HREC/14723) by the Human Research Ethics Committee (HREC) of the University of Abuja Teaching Hospital Gwagwalada (UATH), Abuja, Nigeria.
Authors’ ORCIDs
- Abdullahi IN: https://orcid.org/0000-0002-5511-1272
- Dangana A: https://orcid.org/0000-0001-9955-3841
- Ugwu CN: https://orcid.org/0009-0008-6910-8665
- Akyala AI: https://orcid.org/0000-0002-4168-6104
- Dansura ML: https://orcid.org/0009-0008-1965-5046
- Samuel BE: https://orcid.org/0009-0007-8690-4850
- Gagari VF: https://orcid.org/0009-0003-0460-3888
- Gyang NM: https://orcid.org/0009-0002-0560-8560
- Uzoebo NK: https://orcid.org/0009-0000-8001-1444
Competing interests
The authors declare that they have no competing interests.
References
[1] Ajuwon BI, Yujuico I, Roper K, Richardson A, Sheel M, Lidbury BA. Hepatitis B virus infection in Nigeria: a systematic review and meta-analysis of data published between 2010 and 2019. BMC Infect Dis. 2021 Oct;21(1):1120. DOI: 10.1186/s12879-021-06800-6[2] Egbe KA, Ike A, Egbe F. Knowledge and burden of hepatitis B virus in Nasarawa State, Nigeria. Sci Afr. 2023;22:e01938. DOI: 10.1016/j.sciaf.2023.e01938
[3] Center for Disease Control and Prevention. Clinical Testing and Diagnosis for Hepatitis B. 2023 [last access 2025 Jan 20]. Available from: https://www.cdc.gov/hepatitis-b/hcp/diagnosis-testing/index.html
[4] Olayinka AT, Oyemakinde A, Balogun MS, Ajudua A, Nguku P, Aderinola M, Egwuenu-Oladejo A, Ajisegiri SW, Sha'aibu S, Musa BO, Gidado S, Nasidi A. Seroprevalence of Hepatitis B Infection in Nigeria: A National Survey. Am J Trop Med Hyg. 2016 Oct;95(4):902-7. DOI: 10.4269/ajtmh.15-0874
[5] Musa BM, Bussell S, Borodo MM, Samaila AA, Femi OL. Prevalence of hepatitis B virus infection in Nigeria, 2000-2013: a systematic review and meta-analysis. Niger J Clin Pract. 2015;18(2):163-72. DOI: 10.4103/1119-3077.151035
[6] Tian Y, Xu Y, Zhang Z, Meng Z, Qin L, Lu M, Yang D. The amino Acid residues at positions 120 to 123 are crucial for the antigenicity of hepatitis B surface antigen. J Clin Microbiol. 2007 Sep;45(9):2971-8. DOI: 10.1128/JCM.00508-07
[7] Chen J, Li L, Yin Q, Shen T. A review of epidemiology and clinical relevance of Hepatitis B virus genotypes and subgenotypes. Clin Res Hepatol Gastroenterol. 2023 Aug;47(7):102180. DOI: 10.1016/j.clinre.2023.102180
[8] Dong Z, Li JR, Zhao ZX, Xu L, Yu W, Kang WY, Li QF. Molecular epidemiology of hepatitis B virus genotypes and subgenotypes in ethnic minority populations, Yunnan province, China. Epidemiol Infect. 2021 Nov;150:e11. DOI: 10.1017/S0950268821002326
[9] Kafeero HM, Ndagire D, Ocama P, Kato CD, Wampande E, Walusansa A, Kajumbula H, Kateete D, Ssenku JE, Sendagire H. Mapping hepatitis B virus genotypes on the African continent from 1997 to 2021: a systematic review with meta-analysis. Sci Rep. 2023 Apr;13(1):5723. DOI: 10.1038/s41598-023-32865-1
[10] Liu Z, Zhang Y, Xu M, Li X, Zhang Z. Distribution of hepatitis B virus genotypes and subgenotypes: A meta-analysis. Medicine (Baltimore). 2021 Dec;100(50):e27941. DOI: 10.1097/MD.0000000000027941
[11] Kyaw YY, Lwin AA, Aye KS, Thu HM, Htun MM, Soe HO, Aye KT, Thant KZ, Hwang HJ, Cheong J. Distribution of hepatitis B virus genotypes in the general population of Myanmar via nationwide study. BMC Infect Dis. 2020 Jul;20(1):552. DOI: 10.1186/s12879-020-05269-z
[12] Toyé RM, Loureiro CL, Jaspe RC, Zoulim F, Pujol FH, Chemin I. The Hepatitis B Virus Genotypes E to J: The Overlooked Genotypes. Microorganisms. 2023 Jul;11(8):1908. DOI: 10.3390/microorganisms11081908
[13] Schillie S, Vellozzi C, Reingold A, Harris A, Haber P, Ward JW, Nelson NP. Prevention of Hepatitis B Virus Infection in the United States: Recommendations of the Advisory Committee on Immunization Practices. MMWR Recomm Rep. 2018 Jan;67(1):1-31. DOI: 10.15585/mmwr.rr6701a1
[14] Kumar M, Pahuja S, Khare P, Kumar A. Current Challenges and Future Perspectives of Diagnosis of Hepatitis B Virus. Diagnostics (Basel). 2023 Jan;13(3):368. DOI: 10.3390/diagnostics13030368
[15] Deng X, Guo X, Gu H, Wang D, Laperche S, Allain JP, Zang L, Candotti D. Anti-HBc-nonreactive occult hepatitis B infections with HBV genotypes B and C in vaccinated immunocompetent adults. J Viral Hepat. 2022 Nov;29(11):958-67. DOI: 10.1111/jvh.13733
[16] Lazarevic I, Banko A, Miljanovic D, Cupic M. Clinical Utility of Quantitative HBV Core Antibodies for Solving Diagnostic Dilemmas. Viruses. 2023 Jan;15(2):373. DOI: 10.3390/v15020373
[17] Adoga MP, Gyar SD, Pechulano S, Bashayi OD, Emiasegen SE, Zungwe T, Iperepolu OH, Agupugo C, Agwale SM. Hepatitis B virus infections in apparently healthy urban Nigerians: data from pre-vaccination tests. J Infect Dev Ctries. 2010 Jun;4(6):397-400.
[18] Abazuh UD, Adebayo OH, Losh NB, Adeyemo DA, Bello OK, Oyediran OS, et al. Detection and prevalence of HBsAg and HBV DNA among visiting patients attending health facilities in Nigeria. Sci Afri. 2023;20:e01702. DOI: 10.1016/j.sciaf.2023.e01702
[19] Ingasia LAO, Kostaki EG, Paraskevis D, Kramvis A. Global and regional dispersal patterns of hepatitis B virus genotype E from and in Africa: A full-genome molecular analysis. PLoS One. 2020;15(10):e0240375. DOI: 10.1371/journal.pone.0240375
[20] Gnyawali B, Pusateri A, Nickerson A, Jalil S, Mumtaz K. Epidemiologic and socioeconomic factors impacting hepatitis B virus and related hepatocellular carcinoma. World J Gastroenterol. 2022 Aug;28(29):3793-802. DOI: 10.3748/wjg.v28.i29.3793
[21] Anejo-Okopi J, Okeke E, Davwar PM, Onwuamah C, Onywera H, Omaiye P, Duguru M, Okojokwu OJ, Ujah OI, Jonathan B, George CA, Crown RS, Yakubu FB, Sokei JO, Okoli LC, Audu O, Inzaule SC, Abah IO, Agaba P, Agbaji OO, Sagay AS, Hawkins C. Molecular detection of hepatitis B virus genotype E with immune escape mutations in chronic hepatitis B patients on long-term antiviral therapy in Jos, Nigeria. Afr J Lab Med. 2022;11(1):1677. DOI: 10.4102/ajlm.v11i1.1677
[22] Bukbuk D, Bassi A, Mangoro Z. Sero-prevalence of hepatitis B surface antigen among primary school pupils in rural Hawal valley, Borno State, Nigeria. J Comm Med Primary Health Care. 2005;17(1):20-3. DOI: 10.4314/jcmphc.v17i1.32420
[23] Bello RH, Obot E, Olabode HOK. Sero-prevalence and risk factors associated with hepatitis B surface antigen (HBsAG) amongst patients in BIU, Borno State, Nigeria. J Public Health Epidemiol. 2011;3(10):448-53. Available from: https://academicjournals.org/article/article1381230930_Bello%20et%20al.pdf
[24] Iwalokun BA, Hodonu SO, Olaleye BM, Olabisi OA. Seroprevalence and biochemical features of hepatitis B surface antigenemia in patients with HIV-1 infection in Lagos, Nigeria. Afr J Med Med Sci. 2006 Sep;35(3):337-43.
[25] Nwokedi EO, Odimayo MS, Emokpae AM, Yahaya IA, Sadiq MN, Okwori EE. Seroprevalence of hepatitis B surface antigen among patients attending Aminu Kano Teaching Hospital, Kano. Niger J Med. 2010;19(4):423-6. DOI: 10.4314/njm.v19i4.61968
[26] GBD 2019 Mental Disorders Collaborators. Global, regional, and national burden of 12 mental disorders in 204 countries and territories, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Psychiatry. 2022 Feb;9(2):137-50. DOI: 10.1016/S2215-0366(21)00395-3
[27] Burdette DL, Lazerwith S, Yang J, Chan HLY, Delaney Iv WE, Fletcher SP, Cihlar T, Feierbach B. Ongoing viral replication and production of infectious virus in patients with chronic hepatitis B virus suppressed below the limit of quantitation on long-term nucleos(t)ide therapy. PLoS One. 2022;17(4):e0262516. DOI: 10.1371/journal.pone.0262516
[28] Ahmad AE, Bakari AG, Musa BOP, Mustapha SK, Jamoh BY, Abdullahi IN, Tahir MI, Olatunji AO, Maishanu SH, Suleiman AB, Tolulope A, Hawkins C, Sagay AS, Zoakah A, Olayinka AT. Pattern of prevalent Hepatitis B virus genotypes in Zaria, Nigeria. Niger Postgrad Med J. 2019;26(2):80-6. DOI: 10.4103/npmj.npmj_59_19
[29] Ezea NMC, Chukwuma NOG, Aneke NCC, Ejinaka NP, Nchinda NGW, et al. Pattern and prevalence of hepatitis B virus genotypes in among discordant partners Enugu, Southeastern Nigeria. GSC Biol Pharmacl Sci. 2022;18(2):144-53. DOI: 10.30574/gscbps.2022.18.2.0058
[30] Umego CF, Mboto CI, Asitok AD, Osaji LC, George UE, Edet UO, Mbim EN, Faleye TO, Adewumi OM, Adeniji JA. Circulation of hepatitis B virus genotype-E among outpatients in tertiary hospitals in the Niger-Delta region of Nigeria. Afr Health Sci. 2022 Mar;22(1):511-20. DOI: 10.4314/ahs.v22i1.60
[31] Jose-Abrego A, Roman S, Laguna-Meraz S, Panduro A. Host and HBV Interactions and Their Potential Impact on Clinical Outcomes. Pathogens. 2023 Sep;12(9):1146. DOI: 10.3390/pathogens12091146
[32] Simmonds P, Midgley S. Recombination in the genesis and evolution of hepatitis B virus genotypes. J Virol. 2005 Dec;79(24):15467-76. DOI: 10.1128/JVI.79.24.15467-15476.2005
[33] Anabire NG, Quaye O, Helegbe GK. Circulation of multiple hepatitis B virus genotypes in individual pregnant women seeking antenatal care in northern Ghana. Virol J. 2023 Jul;20(1):149. DOI: 10.1186/s12985-023-02110-2
[34] Naito H, Hayashi S, Abe K. Rapid and specific genotyping system for hepatitis B virus corresponding to six major genotypes by PCR using type-specific primers. J Clin Microbiol. 2001 Jan;39(1):362-4. DOI: 10.1128/JCM.39.1.362-364.2001




