Privacy-preserving record linkage by a federated trusted third party (fTTP) – unlocking medical research potential in Germany
Christopher Hampf 1Martin Bialke 1
Hauke Hund 2
Christian Fegeler 2
Stefan Lang 3
Peter Penndorf 1
Nico Wöller 1
Frank-Michael Moser 4
Arne Blumentritt 4
Ronny Schuldt 4
Florian Seidel 5
Peter Brunecker 6
Reto Wettstein 7
Lukas Arnecke 8
Wolfgang Hoffmann 1
1 Institute for Community Medicine, Department Epidemiology of Health Care and Community Health, University Medicine Greifswald, Germany
2 GECKO Institute, Heilbronn University of Applied Sciences, Heilbronn, Germany
3 Gefyra GmbH, Münster, Germany
4 Trusted Third Party of the University Medicine Greifswald, Germany
5 Berlin Institute of Health at Charité – Universitätsmedizin Berlin, Project HiGHmed, Berlin, Germany
6 Berlin Institute of Health at Charité – Universitätsmedizin Berlin, MeDIC, Berlin, Germany
7 Institute of Medical Informatics, University Hospital Heidelberg, Germany
8 Zentrum für Digitalisierung und Informationstechnologie, University Hospital Heidelberg, Germany
Abstract
Introduction: In the context of the COVID-19 pandemic, the urgent need for centralized research infrastructures became clear in Germany.
Based thereon, large multicenter medical research projects, such as the Network of University Medicine (NUM) were initiated with the aim to collaboratively consolidate and harmonize the medical data of the individual healthcare providers for research purposes.
As a part of the Medical Informatics Initiative (MII), data integration centers (DIC) have already been established at all university hospitals to provide standardized data sets. In order to comply with the relevant data protection requirements, a privacy-preserving record linkage (PPRL) is required to enable cross-site merging of patient records. A federated trusted third party (fTTP) was implemented.
Material and methods: Generic use cases were identified, conceptualized and implemented and provide the basis for the research work. Different scenarios such as rare diseases were considered, with the option to extend the PPRL using additional record linkage methods. Existing tools such as identity and pseudonym management systems were expanded regarding their respective functionalities.
Results: The fTTP enables PPRL and pseudonymization of patient records. It uses well-established PPRL methods. A pseudonymization hierarchy enables the individual’s re-identification e.g., in cases in which incidental findings require contacting a participant in research projects. A clearing procedure is set up to manage potential matches in order to minimize homonym and synonym errors.
Conclusion: The fTTP enables a unified, project-specific pseudonymization across multiple participating sites and PPRL. The fTTP enables secure linkage of further medical data sources, such as data from clinical studies or claims data, on a patient-specific basis. The outlined concepts and technical implementation can serve as blueprint to further use cases.
Keywords
privacy-preserving record linkage, pseudonymization, federated Trusted Third Party, identity management, data quality, data sharing
Introduction
In the context of the COVID-19 pandemic, the importance of medical data’s (MDAT) timely availability provided by clinical healthcare settings to monitor and manage medical care was recognized. Governments worldwide were under pressure to quickly provide recommendations and regulations regarding infection control measures against the spread of the SARS-CoV-2 virus which needed to be regularly adapted based on the respective infection situation. It became obvious that centralized infrastructures were urgently needed to access the existing medical data which is held. They are decentralized by various healthcare providers for research purposes in order to ensure data-based decision-making. The Network of University Medicine (NUM) was initiated in April 2020, whereby all 36 university hospitals in Germany collaborated to establish research data infrastructures in order to monitor the current healthcare situation, to coordinate multicenter research projects and to prepare for future pandemics. Currently, 13 interdisciplinary research projects were initiated to gain rapid and robust findings to answer urgent research questions [1].
However, various challenges arose during the establishing and implementing of centralized (national) research infrastructures in Germany. This include a coherent data format for data collection, heterogeneity for different decentralized hospital information systems, or merging different data sources into one (e.g. electronic health records (EHR), laboratory data, radiological data and pathology reports, which are recorded using varying electronic systems depending of the hospital). Therefore, the Medical Informatics Initiative (MII) established data integration centers (DIC) for each of the 36 university hospitals to harmonize and unite MDAT for research purposes. The NUM CODEX (COVID-19 Data Exchange Platform) project [2] aimed to implement a central COVID-19 research platform to provide data sets in the GECCO83 (German Corona Consensus) [3] format for multicenter research in a patient-related pseudonymized manner [4]. The NUM-RDP (NUM Routine Data Platform) follow-up project [5] was built on the previously established CODEX project’s infrastructures and components of the MII [6] and extended the available data with routine data. Several structural and organizational as well as data protection issues were collaboratively discussed. One key aspect in this context is that one person’s data within different systems and across different data sources is merged in a secure and person-specific manner, as an individual’s medical data are often collected and stored at multiple sites.
Different datasets referring to the same person may appear multiple times in a multicenter study. Local trusted third parties (TTP) were established at each DIC, which match, merge and manage personally identifiable information (PII), manage pseudonyms, manage the patients’ informed consents (IC), and coordinates withdrawal processes if requested by the patient. Beside these decentralized components a federated trusted third party (fTTP) that performs a privacy-preserving record linkage (PPRL) and generates cross-site pseudonyms was needed. The goal is to link a person’s MDAT based on an error-tolerant record linkage method, so that all their data sets can be linked and result in a complete data set. In contrast to countries like the United Kingdom or South Korea [7], in Germany there is no unique identifier available [8] so that PIIs, such as first name, surname, gender, or date of birth are used to determine whether two records belong to the same person. Due to incorrect variations of PIIs, a record linkage can lead to ambiguous matching results. To resolve these cases, a clerical review is conducted, which involves a manual assessment to determine whether two data records belong to the same person. This is important to minimize duplicate records for the same person so-called synonym errors.
To protect data privacy, PPRL approaches allow to link PII belonging to the same person without revealing the attribute values [9]. A well-established method for PPRL is the usage of bloom filters (BF) at the data collecting site [10], [11], [12]. A BF is a bit vector of length n and is initially occupied by n zeros [11]. The BF is a data structure in which information is hashed by switching bit positions to one. In contrast to other hashing methods like MD-5 or SHA1, small differences within the input data result in a similar BF. As a consequence, BFs allow similarity comparisons based on individually encoded attributes so-called field-level BF [13] (e.g. the first name), or based on merged BF of various attributes so-called cryptographic long-term keys (CLK) [12], [14]. To prevent attacks against BF [12], [15], [16], [17], [18] e.g. dictionary or frequency attacks, a combination of random hashing, CLK and additional hardening techniques are preferred [12], [19], [20], [21], [22], [23]. The similarity comparison of BFs is performed by a linkage unit [24]. Regardless of the used BF method, the probability of having the same input data increases the more bit positions align between two BFs. The Tanimoto coefficient may be used as a similarity measure for bit vectors [25].
After record linking it is essential to generate pseudonyms that refer to one person. A pseudonym is – in the best case – a context-free string based on random characters. A pseudonym – in principle – enables a person’s re-identification e.g. to allow a contact in case of incidental findings [26].
Objectives
This publication describes different use cases, initial technical implementation as well as conception of a fTTP exemplified by the German projects CODEX and NUM-RDP based on consolidated general concepts of the MII. The fTTP shall support both a cross-site PPRL with encoded PII in form of BF and, if necessary, a selective PII matching for clerical reviews. Moreover, a core feature is supporting the uniform secondary pseudonymization of MDAT for data transfers.
Material and methods
The NUM-RDP project [5], requires a secure cross-site record linkage in a privacy-preserving manner for medical data matching. The NUM infrastructure provides central components: a fTTP that performs the PPRL and generates cross-site pseudonyms for medical data sets, a data transfer hub (DTH) [27] that transfers MDAT from the DICs to the routine data platform and replaces pseudonyms interactively with the fTTP, and a central routine data platform to provide pseudonymized MDAT to researchers.
The MII developed basic options for federating local TTPs [28]. The main fTTP’s functions are the unified pseudonymization across all connected DICs and a record linkage in a privacy-preserving manner. This results in a PPRL able to assign project-specific, cross-site pseudonyms to one person allowing for linkage between all MDAT collected at multiple sites for the individual. Taking into account the separation principle [29], [30], MDAT and PII are separated and only data required for record linkage is transmitted to the fTTP. Thus, the fTTP receives encoded PII in form of BF.
The TTP’s identity management classifies the compared records as matching, potentially matching or not matching based on the calculated attributes’ similarities [8], [31]. In case of a potential match, a clerical review is often performed manually comparing the matching attributes (e.g. first name, surname, birth date etc.) and classifying the data sets as match or no match [8], [32]. Although PPRL and PII based record linkage methods achieve the same match quality [30], in cases of potential matches, a clerical review is necessary [8]. By design, data correction or clearing is not possible with BF alone. In the cases of potential matches with a BF, a following record linkage using PII may be necessary in particular cases, containing a low number of patients and the aim of minimizing homonym and synonym errors [28]. Therefore, the fTTP is separated into two components: fTTP-probability, for PPRL as default record linkage method and fTTP-clearing, in cases of potential matches with a following project-specific record linkage in cooperation with the respective DIC and a temporary cache of PII, to increase the data quality. The separation of the two components ensures BF’s and PII’s separation and enables the processing at different locations and, if necessary, by different organizations.
The requirements regarding functionalities, infrastructure and desired pseudonym hierarchy are project-specific. The RDP project is used as an example to describe the fTTP.
Identification of use cases and processes
In terms of the concept of decentralization within the MII and a clear separation of the DIC’s responsibilities, the following prerequisites apply:
- DICs are responsible for verifying persons’ consents to ensure that persons permit data transfers of their MDAT for research. Only persons with a valid consent are registered at the fTTP.
- DICs generate the CLK-BF based on PII and the same BF method so that BF can be compared within the fTTP. Only a few types of errors can be detected by the fTTP like different BF lengths.
- PII used in the DICs are complete and correct so that MDAT are only merged based on BF that were generated based on real and correct PII.
The fTTP-probability performs a PPRL and generates, links, and manages site-specific and cross-site pseudonyms. Having site-specific pseudonyms for each patient assures the patient’s previous treatments to be uneducable at other sites. DICs transfer encrypted MDAT as well as their site-specific pseudonym to the DTH. The DTH requests the cross-site pseudonym from the fTTP. The cross-site pseudonym and the encrypted MDAT are sent to the routine data platform. Only the cross-site pseudonym is known to the routine data platform. Subsequently, the MDAT source and institute remain anonymous. Additionally, MDATs are pseudonymized through transfer-specific pseudonyms generated and linked by the fTTP when exporting them to a researcher. De-pseudonymization is possible to re-contact a patient in accordance with the law “Right of access by the data subject” (Article 15 EU GDPR (General Data Protection Regulation)). In these instances, the fTTP needs to cooperate with the DICs as they keep the patient’s PIIs.
The fTTP-clearing performs a record linkage on temporarily cached PII. This process is triggered only when a record linkage based on BF within the fTTP-probability reaches a potential match. For projects with large cohorts this component can be omitted if not needed. Projects containing small numbers of participants (e.g. in the context of rare diseases) may require correct matching, an fTTP-clearing should be added to minimize matching errors.
Results
We identified these four use cases to enable a PPRL, pseudonymization, and clearing within the fTTP:
- Use case 1 (UC1): Person registration based on BF and generating site-specific and cross-site pseudonyms
- Use case 2 (UC2): Re-pseudonymization of site-specific pseudonyms to cross-site pseudonyms
- Use case 3 (UC3): Generating project-specific transfer pseudonyms for data transfers to researchers
- Use case 4 (UC4): Clearing process to resolve potential BF-matches by accessing PII
(UC1) Person registration based on BF and generating of site-specific and cross-site pseudonyms
UC 1 comprises the registration of persons triggered by a local TTP. The fTTP performs a PPRL based on the transferred BF and classifies the result as match, potential match or no match. During the initial registration respectively if the classification is no match, two pseudonyms are generated:
- The site-specific pseudonym that is only disclosed to the site sending the BF.
- The cross-site pseudonym that is only disclosed to the routine data platform and uniquely references a person in the entire project across all sites.
If a person is already known, due to a previous registration by another DIC, the fTTP classifies the BF as match. Solely a site-specific pseudonym will be generated and assigned to the existing cross-site pseudonym. If a site sends the same BF multiple times, the fTTP will always reply with the same site-specific pseudonym. To complete a person’s registration process the fTTP sends a BF-specific response containing the site-specific pseudonym to the registering DIC.
In case of a potential match, UC4 will be triggered as a sub process. The registration process based on BF is summarized in Figure 1 [Fig. 1].
Figure 1: Process to register a person in the fTTP-probability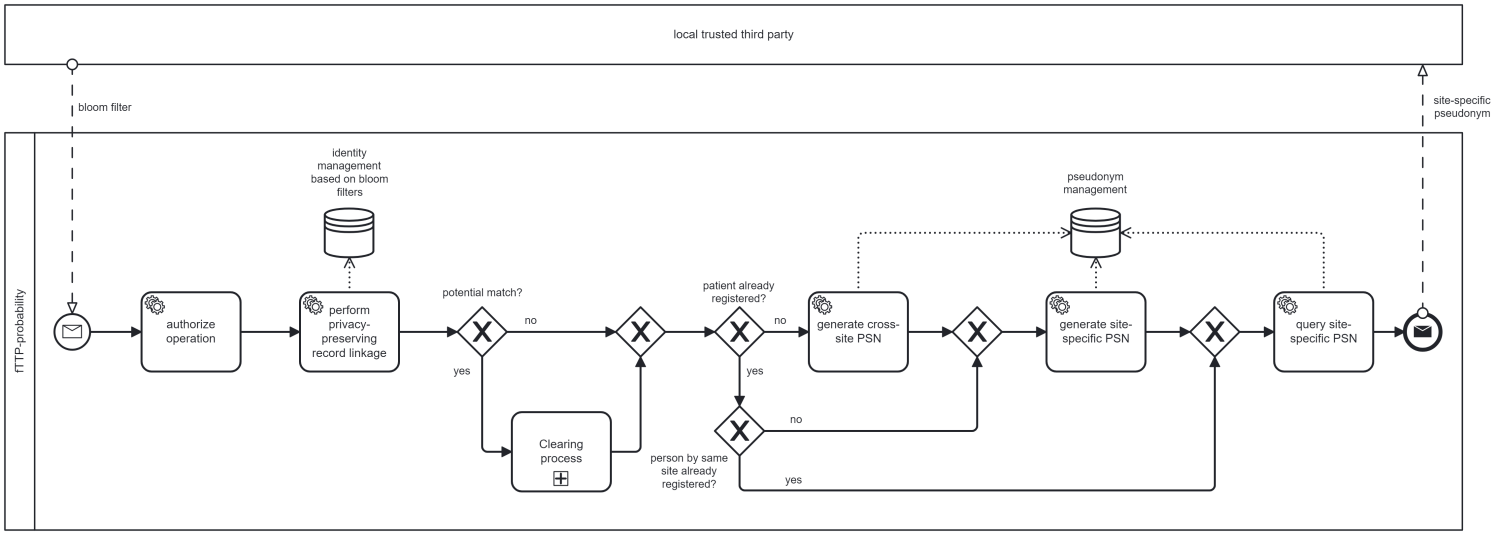
(UC2) Re-Pseudonymization of site-specific pseudonyms to cross-site pseudonyms
If MDAT is transferred from one DIC-site to the routine data platform, UC2 describes the data re-pseudonymization with the fTTP’s support. The DTH initiates this re-pseudonymization by sending site-specific pseudonyms to the fTTP. They validate the pseudonyms and respond with the assigned cross-site pseudonyms. The DTH is responsible for replacing site-specific pseudonyms with cross-site pseudonyms as well as sending MDAT to the routine data plattform. The process is summarized in Figure 2 [Fig. 2].
Figure 2: Process of re-pseudonymization by the DTH interacting with the fTTP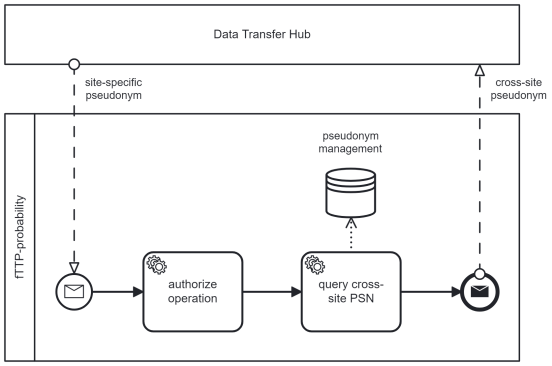
(UC3) Generating project-specific pseudonyms for data transfers to researchers
If the routine data platform gets a request for an MDAT-transfer intended for a specific research project, an additional project-specific pseudonym will be generated. The researchers receive only MDAT containing project-specific pseudonyms for their particular project. First, the routine data platform requests the fTTP with the known cross-site pseudonym. The fTTP generates a new project-specific pseudonym, assigns it to the cross-site pseudonym within the pseudonym hierarchy and forwards the project-specific pseudonym. Based on it, the fTTP can determine which persons were involved in which projects. For incidental findings occurring during a research project which allow for hierarchy also allows to resolve the pseudonyms in cooperation with the registering DIC to re-contact the respective person. The process for generating project-specific pseudonyms is summarised in Figure 3 [Fig. 3].
Figure 3: Process of requesting a project-specific pseudonym by the Routine Data Platform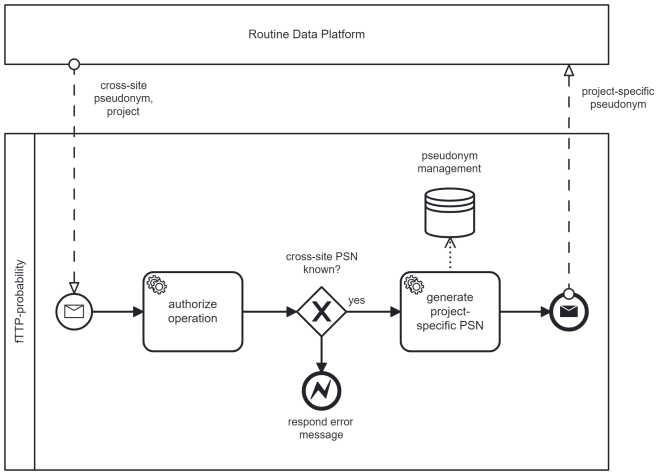
(UC4) Clearing process to resolve potential BF-matches by accessing PII
If UC1 triggers a clearing process, it is executed before pseudonyms are generated and transferred to the requesting DIC. At least two DICs have to be informed: (1) the DIC that sent the BF causing to a potential match and (2) one or more DICs that previously registered a BF that is now a potential match. (1) Instead of a site-specific pseudonym, an information about the potential match will be sent so that the DIC can send PIIs to the fTTP-clearing. The DIC can periodically request a task system to realize this. For (2), DICs are informed of a potential match by their respectively known site-specific pseudonyms. These DICs then send the corresponding PII to the site-specific pseudonyms. In both cases, the DICs receive a list of requested project-specific PIIs (e.g. first name, surname, birth date, etc.).
When all involved DICs have sent PIIs to the fTTP-clearing a record linkage based on PIIs is processed. This is an automatic process, but in case of potential match (based on PII) may require clerical review. The DIC’s consultation may be necessary for data correction or additional data provision. After the (automatic or manual) classification into match or no match, the fTTP-clearing deletes the cached PIIs and sends the result to the fTTP-probability, which in turn generates pseudonyms. The DIC, whose BF triggered the clearing process, obtains the site-specific pseudonym. This process is depicted in Figure 4 [Fig. 4].
Figure 4: Process of clearing bloom filter based potential matches by the fTTP-clearing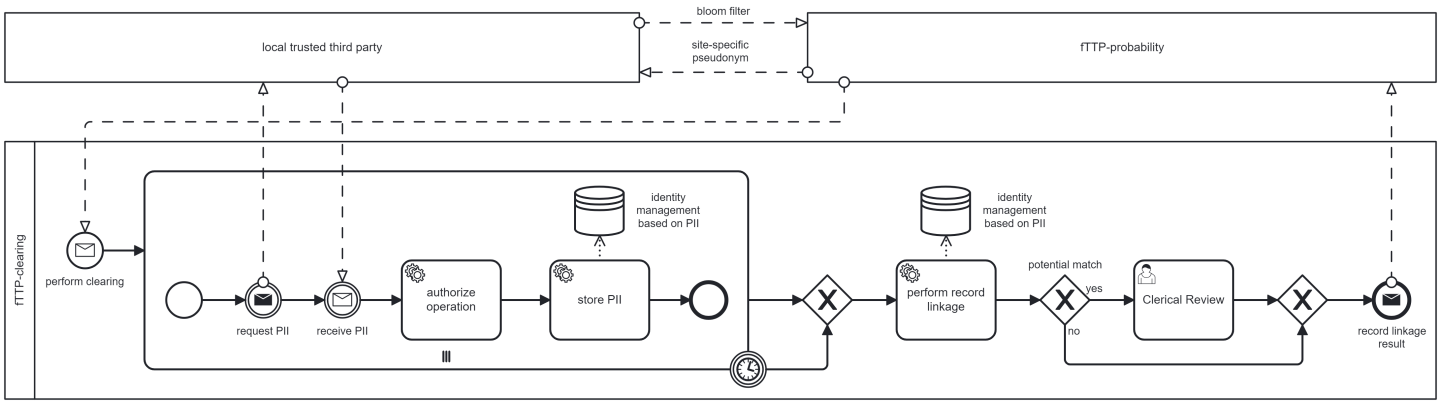
Technical implementation
The fTTP’s technical infrastructure setup is in accordance with the recommended measures for segmentating networks by the Federal Office for Information Security (BSI) [33]. This includes a demilitarised zone (DMZ), which implements the site’s authentication by using a client certificate. In addition, access is only granted for approved IPs or via a site-specific login. Connections are routed through an additional internal transfer zone. Applied tools for record linkage, pseudonym management, and workflow realization etc. are processed within a separate trustee zone. The components of fTTP-probability and fTTP-clearing are performed by different logical machines, subsequencly PIIs and BFs are never on the same logical system. Each DIC receives a site-specific API key that only allows access to DIC permitted functions. For example, the DTH may only perform re-pseudonymization, and a local TTP may only register persons. This concept fullfills the requirements of large national projects like the German Centre for Cardiovascular Research (DZHK) [34] or German National Cohort (NAKO) [35] working with PIIs, however, the fTTP receives only BFs in most cases.
The technical interfaces were specified in Health Level 7 (HL7) Fast Healthcare Interoperability Resources (FHIR®) as required by MII and NUM. To ensure FHIR® conformity, the company Gefyra (https://gefyra.info/) was commissioned to support the specification process to validate corresponding implementations and to propose corrections if applicable. The interfaces were continuously documented in the public Simplifier project of the Independent Trusted Third Party of the University Medicine Greifswald [36]. A public ballot for comments according to the HL7 guidelines was successfully processed as well as all community comments, including stakeholders from all four MII consortia.
A technical connection can be established through the provided REST-interface. Beside other frameworks in NUM and MII the wide-spread data sharing framework (DSF) [27], [37] allows for a direct connection to the fTTP. Therefore, most of the DICs do not require a proprietary implementation of the fTTP interfaces.
The technical implementation is based upon well established TTP-tools developed by the Independent Trusted Third Party of the University Medicine Greifswald. The implementation of the FHIR interfaces was realized by the Trusted Third Party FHIR Gateway (TTP-FHIR Gateway) using the HAPI FHIR imlementation for HL7 FHIR [38], [39]. This FHIR®-specific, supplementary module coordinates and validates incoming FHIR® requests and forwards them to the assigned fTTP-components. Specific processes are modeled using the workflow management component TTP Dispatcher [40]. It allows a connection to the identity management system (E-PIX® [28], [31]) that performs PPRL as well as the pseudonym management system (gPAS®) to generate pseudonyms and implement pseudonym hierarchies.
E-PIX® was extended by functionalities for generating and similarity-based matching of BFs. Thus, local TTPs are able to generate the required BFs with a local instance of E-PIX®. The fTTP is able to match BFs, therefore, E-PIX® is used within the fTTP as well. E-PIX® was published for public use.
All these tools are available for interested sites as they are open source (https://github.com/mosaic-hgw/). This allows a fast and easy way to implement the technical components that allow a connection to large national research networks like the MII and NUM. Extending the networks i.e. including more institutions, e.g. hospitals and major private practices as sources of medical date, poses a prospective challenge for the future.
Discussion
Bloom filter among other PPRL techniques
BFs are a popular method for PPRL. Other PPRL techniques like secure multi-party computation (SMPC) are provably secure, however, they are costly in regards of communications and computation [23], [24]. Due to the need of a quick and easy solution, BFs were chosen as a well-established method for PPRL. In combination with state of the art methods and hardening mechanisms, BFs are a secure solution. Attackers, besides overcoming security mechanisms, must prepare highly to gain access to BFs [17]. In addition, the most prevalent local identity management solutions used by the DICs in NUM and MII already have implemented BF methods. Hence, they use existing tools for BF instead of establishing a new infrastructure for SMPC. Finally, SMPC is not a replacement for an fTTP as it is an organizational unit with responsibilities beyond record linkage to transparently and responsibly address legal and technical requirements, such as right of access and standardized pseudonymization.
Cross-site record linkage methods based on PII are well-implemented in multicenter research infrastructures like the DZHK or the NAKO. There, potential matches range between 1.3% and 7.2%, which were manually resolved through clerical review [41]. Based on the fact that BFs lead to the same matching results as unencrypted PIIs (33), it is assumed that a similar number of potential matches occur.
At the end of March 2021, a first demonstrator event showed the correct implementation of UC1 and UC2 with the help of the Charité – University Medicine Berlin, the University Hospital Heidelberg and the DTH, which is being operated at Heilbronn University [42]. All 34 DICs of NUM-RDP implemented a validated connection to the fTTP since November 2022.
Supporting orchestration of consent and withdrawal processes
A patient’s valid informed consent (based on the MII broad consent) is the legal basis for processing and scientific use of routine health-care data in NUM. Currently, the patient’s consent is locally documented and processed at the university hospital’s local TTPs. In order to be able to implement patients’ data protection rights in conformity with the GDPR, the central platform of RDP may only provide research data for scientific analyses if the validity of the informed consent has been unequivocally ensured by the NUM-RDP project. A federated consent management would be an expedient extension to the fTTP-concept for orchestrating cross-site consents and withdrawal processes in order to support the correct and timely implementation of data subjects rights at the NUM sites and to the RDP project in a transparent, consistent and uniform manner. Currently, limited MDAT, i.e. only a few hundred persons, were transmitted to the routine data platform. The reasons for this include the fact that the establishment of the required informed consent was delayed by local ethics committees’ disapproval. Some sites still recruit only small numbers of participants. Therefore, so far there were no potential matches, and no clearing has been necessary so far.
For the first time, a foundation for multi-center use of medical data has been established for research purposes within the framework of NUM by all university hospitals in Germany. This can be prospectively expanded for unexpected events like pandemics. An objective is to extend the networks by including more institutions e.g. local hospitals and private practices as sources for medical data. Synthesizing on other project’s experience with more involved institutions (e.g. DZHK), we presume a similar scalability of the fTTP. The fTTP and PPRL concepts presented here can also serve as a blueprint for further secure intersectoral multicenter research projects involving the linkage of medical data from different data sources. The principles and use cases outlined in this article are in general transferable. The tools used for record linkage and pseudonymization are available as open-source tools. Each university hospital’s DIC was needed due to the heterogeneous data acquisition systems in Germany. On the one hand, it further standardizes data formats and, on the other hand, harmonizes the different data acquisition systems at the hospital level.
Limitations
A decentralized infrastructure is based on distributed responsibilities and trusts that each site will fulfill its due diligence. The fTTP is not able to validate collected informed consents. It must, therefore, rely on the fact that only BF and PII from consented persons are transmitted and withdrawals are communicated promptly. Automatic processes can reduce efforts and delays in communication.
The clerical review within the fTTP-clearing can improve the linkage quality and help to detect errors within the data. In this case requesting PIIs from sites that transferred BFs for potential matches is required. Only available PIIs, which are often used to generate BFs, can be requested. In other contexts, such as cancer registries, additional data sources (e.g. civil registers) are already implemented to resolve potential matches correctly. Therefore, only potential matches with obvious errors within PII can be resolved. In addition to the technical connection, the integration of other data sources also requires a legal evaluation, especially if data come from multiple federal states.
Conclusion
The federated trusted third party was technically designed and successfully implemented into NUM CODEX/RDP projects based on consolidated MII concepts. The initial MII concepts were further developed in collaboration with local trusted third parties and data transfer hub as partners. They were also consolidated and specified by FHIR means for all practical purposes implemented using HAPI FHIR. As of right now, all NUM DICs are connected. In the future, various existing and new NUM projects will be connected to the NUM-RDP infrastructure in order to provide further data for medical research from various data sources. The concept and technical implementation outlined here can serve as a blueprint for further multicenter research initiatives requiring a secure and person-specific linkage of different data sources.
Abbreviations
- BF: bloom filter
- BMBF: Federal Ministry of Education and Research
- BSI: Bundesamt für Sicherheit in der Informationstechnik, Federal Office for Information Security
- CLK: cryptographic long-term key
- CODEX: COVID 19 Data Exchange Platform
- DTH: data transfer hub
- DIC: data integration center
- DZHK: German Centre for Cardiovascular Research
- E-PIX®: enterprise identifier cross-referencing
- FHIR®: Fast Healthcare Interoperability Resources
- fTTP: federated trusted third party
- GECCO: German Corona Consensus
- GDPR: General Data Protection Regulation
- gPAS®: Generic Pseudonym Administration Service
- KKR-MV: Clinical Cancer Registry Mecklenburg-Vorpommern
- NAKO: German National Cohort
- HL7 FHIR®: Health Level 7 Fast Healthcare Interoperability Resources
- IC: informed consent
- IP: internet protocol
- MDAT: medical data
- MII: Medical Informatics Initiative
- NUM: Network of University Medicine
- NUM-RDP: NUM Routine Data Platform
- PII: personally identifiable information
- PPRL: privacy-preserving record linkage
- PSN: pseudonym
- SMPC: secure multi-party computation
- TTP: trusted third party
- TTP-FHIR Gateway: Trusted Third Party FHIR Gateway
- UC: use case
Notes
Availability of data and material
The referenced FHIR documentation is available from https://www.ths-greifswald.de/fttp/fhir/ig/stable. The referenced tools for identity and pseudonym management are available from https://www.ths-greifswald.de/en/researchers-general-public/e-pix/ and https://www.ths-greifswald.de/en/researchers-general-public/gpas/.
Funding
The NUM-project is funded by the Federal Ministry of Education and Research (BMBF) (Grant Number 01KX2021) and MIRACUM (German Federal Ministry of Education and Research, grant number 01ZZ1801M).
Competing interests
In the interest of transparency, it is disclosed that Gefyra GmbH was engaged as a consultant. Additionally, Gefyra GmbH was responsible for the implementation of FHIR profiles and received financial compensation for their services. Beyond that, the authors declare that they have no competing interests.
Authors’ contributions
Christopher Hampf and Martin Bialke contributed equally to this work.
- Drafting of the manuscript: C. Hampf, M. Bialke.
- Expansion of E-PIX®: C. Hampf, F. Moser.
- Harmonisation of interfaces: C. Hampf, H. Hund, M. Bialke.
- Concept and implementation of the TTP-FHIR Gateway: M. Bialke, P. Penndorf.
- Implementation of fTTP Infrastructure: N. Wöller, C. Hampf.
- Participation for demonstration: C. Fegeler, H. Hund, P. Brunecker, F. Seidel, R. Wettstein, L. Arnecke.
- Dispatcher customization: P. Penndorf.
- FHIR profiling and implementational consulting: S. Lang.
- Revision of the manuscript: C. Hampf, M. Bialke, H. Hund, C. Fegeler, S. Lang, P. Penndorf, N. Wöller, F. Moser, A. Blumentritt, R. Schuldt, F. Seidel, P. Brunecker, R. Wettstein, L. Arnecke, W. Hoffmann. All authors approved the final version of the manuscript.
- The figures were created by C. Hampf.
Authors’ ORCIDs
- Christopher Hampf: 0000-0002-4557-4783
- Martin Bialke: 0000-0001-6888-9086
Acknowledgements
Thomas Bahls and Lars Geidel have been involved in projects at the Trusted Third Party of the University Medicine Greifswald for many years and have contributed a large number of ideas and their expertise to this research area.
References
[1] Netzwerk Universitätsmedizin. NUM Projekte. [cited 2023 Sep 26]. Available from: https://www.netzwerk-universitaetsmedizin.de/projekte[2] Prokosch HU, Bahls T, Bialke M, Eils J, Fegeler C, Gruendner J, Haarbrandt B, Hampf C, Hoffmann W, Hund H, Kampf M, Kapsner LA, Kasprzak P, Kohlbacher O, Krefting D, Mang JM, Marschollek M, Mate S, Müller A, Prasser F, Sass J, Semler S, Stenzhorn H, Thun S, Zenker S, Eils R. The COVID-19 Data Exchange Platform of the German University Medicine. Stud Health Technol Inform. 2022 May;294:674-8. DOI: 10.3233/SHTI220554
[3] Sass J, Bartschke A, Lehne M, Essenwanger A, Rinaldi E, Rudolph S, Heitmann KU, Vehreschild JJ, von Kalle C, Thun S. The German Corona Consensus Dataset (GECCO): a standardized dataset for COVID-19 research in university medicine and beyond. BMC Med Inform Decis Mak. 2020 Dec;20(1):341. DOI: 10.1186/s12911-020-01374-w
[4] Netzwerk Universitätsmedizin. CODEX – COVID-19 Data Exchange Platform. [cited 2023 Sep 26]. Available from: https://www.netzwerk-universitaetsmedizin.de/projekte/codex
[5] Heyder R; NUM Coordination Office; NUKLEUS Study Group; NUM-RDP Coordination; RACOON Coordination; AKTIN Coordination; GenSurv Study Group. Das Netzwerk Universitätsmedizin: Technisch-organisatorische Ansätze für Forschungsdatenplattformen [The German Network of University Medicine: technical and organizational approaches for research data platforms]. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2023 Feb;66(2):114-25. DOI: 10.1007/s00103-022-03649-1
[6] Semler SC, Wissing F, Heyder R. German Medical Informatics Initiative. Methods Inf Med. 2018 Jul;57(S 01):e50-e56. DOI: 10.3414/ME18-03-0003
[7] Mills S, Lee JK, Rassekh BM, Zorko Kodelja M, Bae G, Kang M, Pannarunothai S, Kijsanayotin B. Unique health identifiers for universal health coverage. J Health Popul Nutr. 2019 Oct;38(Suppl 1):22. DOI: 10.1186/s41043-019-0180-6
[8] Christen P. Data Matching: Concepts and Techniques for Record Linkage, Entity Resolution, and Duplicate Detection. Heidelberg, Berlin: Springer; 2012.
[9] Vatsalan D, Karapiperis D, Verykios VS. Privacy-Preserving Record Linkage. In: Sakr S, Zomaya A, editors. Encyclopedia of Big Data Technologies. Cham: Springer International Publishing; 2018. p. 1-10. DOI: 10.1007/978-3-319-63962-8_17-2
[10] Bloom BH. Space/time trade-offs in hash coding with allowable errors. Commun ACM. 1970 Jul 13;13(7):422-6.
[11] Schnell R, Bachteler T, Reiher J. Privacy-preserving record linkage using Bloom filters. BMC Med Inform Decis Mak. 2009 Aug;9:41. DOI: 10.1186/1472-6947-9-41
[12] Schnell R, Borgs C. Randomized Response and Balanced Bloom Filters for Privacy Preserving Record Linkage. In: 2016 IEEE 16th International Conference on Data Mining Workshops (ICDMW); 2016 Dec 12-15; Barcelona, Spain. IEEE; 2016. p. 218-24. DOI: 10.1109/ICDMW.2016.0038
[13] Schnell R. Privacy-preserving Record Linkage. In: Harron K, Goldstein H, Dibben C, editors. Methodological Developments in Data Linkage. John Wiley & Sons; 2016. (Wiley Series in Probability and Statistics). p. 201-25.
[14] Schnell R, Bachteler T, Reiher J. A Novel Error-Tolerant Anonymous Linking Code. (German Record Linkage Center (GRLC) Working Paper Series; wp-grlc-2011-02). SSRN; 2011 Nov 16. DOI: 10.2139/ssrn.3549247
[15] Domingo-Ferrer J, Muralidhar K. New directions in anonymization: permutation paradigm, verifiability by subjects and intruders, transparency to users. Inf Sci. 2016 Apr 10;337:11-24.
[16] Kuzu M, Kantarcioglu M, Durham E, Malin B. A Constraint Satisfaction Cryptanalysis of Bloom Filters in Private Record Linkage. Berlin, Heidelberg: Springer; 2011. p. 226-45.
[17] Niedermeyer F, Steinmetzer S, Kroll M, Schnell R. Cryptanalysis of Basic Bloom Filters Used for Privacy Preserving Record Linkage. J Priv Confid. 2014;6(2):59-79. DOI: 10.29012/jpc.v6i2.640
[18] Kroll M, Steinmetzer S. Automated Cryptanalysis of Bloom Filter Encryptions of Health Records (Contribution to the 8th International Conference on Health Informatics, Lisbon, 2015). ArXiv. 2014 Oct 24. DOI: 10.48550/arXiv.1410.6739
[19] Christen P, Vidanage A, Ranbaduge T, Schnell R. Pattern-Mining Based Cryptanalysis of Bloom Filters for Privacy-Preserving Record Linkage. In: Proceedings, Part III. Advances in Knowledge Discovery and Data Mining: 22nd Pacific-Asia Conference (PAKDD 2018); 2018 Jun 3-6; Melbourne, Australia. ACM; 2018. p. 530-42. DOI: 10.1007/978-3-319-93040-4_42
[20] Christen P, Schnell R, Vatsalan D, Ranbaduge T. Efficient cryptanalysis of bloom filters for privacy-preserving record linkage. In: 21st Pacific-Asia Conference on Knowledge Discovery and Data Mining (PAKDD 2017); 2017 May 23-26; Jeju, Korea. Cham: Springer; 2017. (Lecture Notes in Computer Science; 10234). p. 628-40. DOI: 10.1007/978-3-319-57454-7_49
[21] Schnell R, Borgs C. XOR-Folding for hardening Bloom Filter-based Encryptions for Privacy-preserving Record Linkage. (German Record Linkage Center (GRLC) Working Paper Series; WP-GRLC-2016-03). SSRN; 2016 Dec 22. DOI: 10.2139/ssrn.3527984
[22] Franke M, Sehili Z, Rohde F, Rahm E. Evaluation of Hardening Techniques for Privacy-Preserving Record Linkage. In: Proceedings of the 24th International Conference on Extending Database Technology (EDBT); 2021 Mar 23-26; Nicosia, Cyprus. OpenProceedings; 2021. p. 289-300. DOI: 10.5441/002/edbt.2021.26
[23] Heng Y, Armknecht F, Chen Y, Schnell R. On the effectiveness of graph matching attacks against privacy-preserving record linkage. PLoS One. 2022 Sep 22;17(9):e0267893. DOI: 10.1371/journal.pone.0267893
[24] Christen P, Ranbaduge T, Schnell R. Linking Sensitive Data: Methods and Techniques for Practical Privacy-Preserving Information Sharing. Cham: Springer; 2021.
[25] Tanimoto TT. An Elementary Mathematical Theory of Classification and Prediction. New York: International Buisness Machines Corporation; 1958.
[26] Pommerening K, Helbing K, Ganslandt T, Drepper J, Goltz U, Magnor M. Identitätsmanagement für Patienten in medizinischen Forschungsverbünden. Bonn; 2012.
[27] Hund H, Wettstein R, Hampf C, Bialke M, Kurscheidt M, Schweizer ST, Zilske C, Mödinger S, Fegeler C. No Transfer Without Validation: A Data Sharing Framework Use Case. Stud Health Technol Inform. 2023 May;302:68-72. DOI: 10.3233/SHTI230066
[28] Hampf C, Bahls T, Hund H, Drepper J, Lablans M, Speer R. Record Linkage: Optionen für standortübergreifende Datenzusammenführungen. mdi – Forum der Medizin, Dokumentation und Medizin-Informatik. 2019;21(4):117-21.
[29] Boyd JH, Ferrante AM, O'Keefe CM, Bass AJ, Randall SM, Semmens JB. Data linkage infrastructure for cross-jurisdictional health-related research in Australia. BMC Health Serv Res. 2012 Dec;12:480. DOI: 10.1186/1472-6963-12-480
[30] Randall SM, Ferrante AM, Boyd JH, Bauer JK, Semmens JB. Privacy-preserving record linkage on large real world datasets. J Biomed Inform. 2014 Aug;50:205-12. DOI: 10.1016/j.jbi.2013.12.003
[31] Hampf C, Geidel L, Zerbe N, Bialke M, Stahl D, Blumentritt A, Bahls T, Hufnagl P, Hoffmann W. Assessment of scalability and performance of the record linkage tool E-PIX in managing multi-million patients in research projects at a large university hospital in Germany. J Transl Med. 2020 Feb;18(1):86. DOI: 10.1186/s12967-020-02257-4
[32] Randall SM, Ferrante AM, Boyd JH, Semmens JB. The effect of data cleaning on record linkage quality. BMC Med Inform Decis Mak. 2013 Jun 5;13:64. DOI: 10.1186/1472-6947-13-64
[33] Bundesamt für Sicherheit in der Informationstechnik. IT-Grundschutz-Kompendium. 6th ed. Köln: Reguvis; 2023.
[34] Schwaneberg T, Weitmann K, Dösch A, Seyler C, Bahls T, Geidel L, Stahl D, Lee M, Kraus M, Katus HA, Hoffmann W. Data privacy management and data quality monitoring in the German Centre for Cardiovascular Research’s multicentre TranslatiOnal Registry for CardiomyopatHies (DZHK-TORCH). ESC Heart Fail. 2017 Nov;4(4):440-7. DOI: 10.1002/ehf2.12168
[35] German National Cohort (GNC) Consortium. The German National Cohort: aims, study design and organization. Eur J Epidemiol. 2014 May 20;29(5):371-82. DOI: 10.1007/s10654-014-9890-7
[36] Independent Trusted Third Party of the University Medicine Greifswald. IG TTP-FHIR Gateway. [cited 2023 Sep 26]. Available from: https://www.ths-greifswald.de/fttp/fhir/ig/stable
[37] Hund H, Wettstein R, Heidt CM, Fegeler C. Executing Distributed Healthcare and Research Processes - The HiGHmed Data Sharing Framework. Stud Health Technol Inform. 2021 May;278:126-33. DOI: 10.3233/SHTI210060
[38] Smile CDR Inc. HAPI FHIR – The Open Source FHIR API for Java. [cited 2023 Sep 26]. Available from: https://hapifhir.io/
[39] Bialke M, Geidel L, Hampf C, Blumentritt A, Penndorf P, Schuldt R, Moser FM, Lang S, Werner P, Stäubert S, Hund H, Albashiti F, Gührer J, Prokosch HU, Bahls T, Hoffmann W. A FHIR has been lit on gICS: facilitating the standardised exchange of informed consent in a large network of university medicine. BMC Med Inform Decis Mak. 2022 Dec 19;22(1):335. DOI: 10.1186/s12911-022-02081-4
[40] Bialke M, Penndorf P, Wegner T, Bahls T, Havemann C, Piegsa J, Hoffmann W. A workflow-driven approach to integrate generic software modules in a Trusted Third Party. J Transl Med. 2015 Jun 4;13:176. DOI: 10.1186/s12967-015-0545-6
[41] Intemann T, Kaulke K, Kipker DK, Lettieri V, Stallmann C, Schmidt CO, Geidel L, Bialke M, Hampf C, Stahl D, Lablans M, Rohde F, Franke M, Kraywinkel K, Kieschke J, Bartholomäus S, Näher AF, Tremper G, Lambarki M, March S, Prasser F, Haber AC, Johannes Drepper, Schlünder I, Kirsten T, Pigeot I, Sax U, Buchner B, Ahrens W, Semler SC. White Paper – Verbesserung des Record Linkage für die Gesundheitsforschung in Deutschland. Köln: nfdi4health; 2023. DOI: 10.4126/FRL01-006461895
[42] Independent Trusted Third Party of the University Medicine Greifswald. Erfolgreiche Live Demo der föderierten Treuhandstelle (fTTP) in NUM-CODEX. [cited 2023 Sep 26]. Available from: https://www.ths-greifswald.de/erfolgreiche-live-demo-der-foederierten-treuhandstelle-fttp-in-num-codex/




