[Design und Entwicklung von Primern für den Nachweis von Streptococcus pneumoniae, Haemophilus influenzae und Neisseria meningitidis]
Leila Azimi 1Fatemeh Shirkavand 1
Shahnaz Armin 1
Fereshteh Karbasian 2
Hannan Khodaei 1
1 Pediatric Infections Research Center, Research Institute for Children’s Health, Shahid Beheshti University of Medical Sciences, Tehran, Iran
2 Department of pediatric gastroenterology and hepatology, Ali-Asghar children’s hospital, Iran University of Medical Sciences, Tehran, Iran
Zusammenfassung
Hintergrund: Die Sterblichkeitsrate bei Meningitis ist in bestimmten Regionen der Welt immer noch alarmierend hoch. Ziel der Arbeit ist es, die wirksamsten Primer für den Nachweis von Streptococcus (S.) pneumoniae, Haemophilus (H.) influenzae und Neisseria (N.) meningitidis mithilfe der Real-Time-PCR-Technologie zu ermitteln.
Material und Methoden: Es wurden zwei Primer-Sets zum Nachweis von S. pneumoniae, H. influenzae und N. meningitidis unter Verwendung der Primer Biosoft Allele ID 7.6 entwickelt. Untersucht wurden die minimalen bakteriellen Kopienzahlen, die von jedem Primer nachgewiesen werden können, sowie ihre Spezifität.
Ergebnisse: CtrA und hpd2 konnten 400 Kopienzahlen/ml von H. influenzae und N. meningitidis und LytA2 40 Kopienzahlen/ml von S. pneumoniae nachweisen. Die Sensitivität und Spezifität aller Primer betrug 100 % (CI: 95 %).
Schlussfolgerung: Die Verwendung empfindlicherer Primer zum Nachweis des für die bakterielle Meningitis verantwortlichen Erregers erhöht die Chance, die verursachenden Bakterien zu identifizieren. Die entwickelten Primer konnten die ausgewählten Bakterien mit mindestens zehnmal höherer Empfindlichkeit identifizieren als kommerzielle Diagnosekits im Iran.
Schlüsselwörter
Streptococcus pneumoniae, Haemophilus influenzae, Neisseria meningitidis, primer, sensitivity
Introduction
H. influenzae, N. meningitidis, and S. pneumoniae can cause infection in important tissues such as blood and cerebrospinal fluid (CSF), requiring rapid intervention and healing [1], [2], [3], [4], [5]. Fast and early diagnosis of these bacteria is necessary for quick treatment and appropriate antibiotic therapy. Blood and CSF cultures (conventional culture or BACTEC) are traditional methods used to identify bacteria [6]. However, these methods are time-consuming and, in some cases, bacteria cannot grow in the culture medium due to the use of antibiotics and despite the clinical symptoms, bacterial growth is not observed. Molecular methods can be helpful in overcoming these disadvantages.
Real-time PCR, a molecular method, can be used for early diagnosis and fast treatment of the patient. Many commercial diagnosis kits with different levels of sensitivity are used worldwide to identify the cause of infections. The sensitivity and minimum detectable copy numbers of bacteria are related to the primers of the kits. As such, the primers play a key role in these diagnosis kits. In this regard, the aim of this study was to design a diagnostic panel using real-time PCR to identify H. influenzae, N. meningitidis, and S. pneumoniae.
Materials and methods
Samples and setting
H. influenzae, N. meningitidis, and S. pneumoniae, which were isolated from bacterial meningitides and had their identity confirmed by real-time PCR in the last study by our group [7], were selected. These strains were used to evaluate the designed primers.
Primer design
Two different pairs of primers were designed by Allele ID 7.6 for different genes or different parts of genes to detect H. influenzae (hpd gene), N. meningitidis (CtrA and SodC), and S. pneumoniae (lytA) (Table 1 [Tab. 1]).
Table 1: Primer sequencing of specific real-time PCR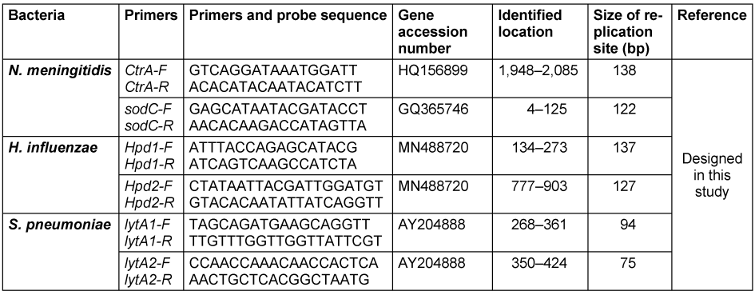
DNA extraction
Bacterial genome extraction was performed using a DNA extraction kit (Qiagen Cat No./ID: 51304). The DNA concentration was read by Qubit. 10 dilutions were prepared by a factor of 101 for each bacterium, and the DNA concentration of all dilutions was determined by the Qbit instrument to determine the sensitivity and the cut-off point of these primers.
Real-time PCR assay
Real time-PCR assay was performed for the identification of bacteria using differently designed primers. Cyber green master mix and ABI step-one, in addition to the real-time PCR instrument, were used in this setup.
The DNA copy number calculator works according to the following equation:
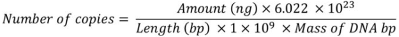
where Amount (ng) is the amount of DNA in nanograms (ng) in the tube, 6.022x1.023 is Avogadro’s constant and represents the number of molecules per mole, Length (bp) is the length of DNA, in base pairs (bp), in the template (H. influenzae 1.830.137 bp, N. meningitidis 2.184.406 bp and S. pneumoniae 2.160.837 bp), 1x109 is the factor used to convert to ng, Mass of DNA bp stands for the average mass of a DNA bp, which is either 660 (dsDNA) or 330 (ssDNA) g/mole. This value depends on what is selected as the type of DNA in the calculator (https://toptipbio.com/dna-copy-number-calculator/).
The genomes of Acinetobacter baumannii, Pseudomonsa (P.) aeruginosa, Escherichia (E.) coli, Klebsiella (K.) pneumoniae, Enterobacter spp., Staphylococcus (S.) aureus and Enterococcus spp. were used as a negative control to evaluate the specificity of these primers.
Results
The results of real-time PCR showing the sensitivity of these primers to detect the included bacteria are presented in Table 2 [Tab. 2].
Table 2: Sensitivity of designed primers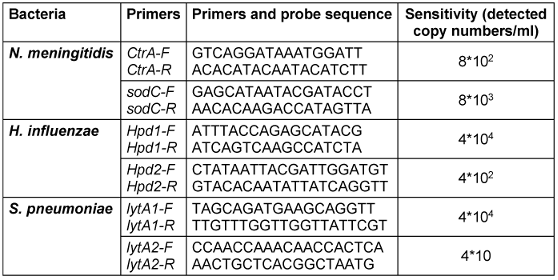
None of the duplication observed with the genomes of A. baumannii, P. aeruginosa, E. coli, K. pneumoniae, Enterobacter spp., S. aureus and Enterococcus spp. confirmed the 100% (CI: 95%) specificity of these primers.
Discussion
Bacterial meningitis occurs most often in childhood and the etiological pathogens can be diverse in different age groups of children [4], [8], [9]. Based on previous studies, the incidence of bacterial meningitis can vary depending on factors such as time, geographical location, and patient age [4], [8], [9].
A systematic review and meta-analysis on the worldwide etiology of bacterial meningitis showed that the most prevalent causative pathogens were N. meningitidis and S. pneumoniae in all age groups, while S. pneumoniae was most prevalent in children [4]. An accurate method to identify these bacteria can be helpful in saving human lives and prevent them from becoming disabled in the future due to meningitis. The results of this study showed higher sensitivity of CtrA primer than SodC for the detection of N. meningitidis. On the other hand, different genome regions of hpd and LytA genes in H. influenzae and S. pneumoniae, respectively, were selected to design primers. The different results when various genes are selected to identify the bacteria, showing that the primer selected for nucleotide sequence between 777–903 has greater sensitivity for detecting H. influenzae. Also, primer design for nucleotide sequences between 350–424 is more sensitive for detecting S. pneumoniae.
In the study by Haddad-Boubaker et al. the results showed that Real-time PCR could detect up to 67.10–4 ng/µL DNA for S. pneumoniae, 38.10–6 ng/µL and 38.10–3 ng/µL for N. meningitidis ctrA gene and sodC gene, respectively, and 97.10–4 ng/µL for H. influenzae [10]. In the current study, the primers used were ctrA, hpd.2, and LytA.2. These results are near to ours.
Cyber green was used in the current study and is more affordable in comparison to using the probe of the Haddad-Boubaker et al. study [10]. One of the commercial molecular-diagnosis kits for the detection of these three bacteria is being used (Sacace™ NHS Meningitidis Real-TM) in Iran. The analytical sensitivity, genome equivalents/ml of this kit is 1*103 (genome of bacteria/ml) [11], but the primers we designed have a minimum detectable bacterial genome 4*102 for N. meningitidis and H. influenzae and 4*10 for S. pneumoniae. These results showed that the primers designed in the current study detected bacteria with at least 10 times greater sensitivity. This is an important advantage of molecular diagnosis kits, especially when patients used antibiotics before the test.
Conclusions
The detection of the causative bacteria of meningitis can be helpful to choose the best therapeutic process as soon as possible. In this regard, selecting the most accurate and rapid method with the greatest possible sensitivity to identify relevant agents is necessary. It is important to know the exact cause of meningitis, because the choice of treatment depends on it.
Notes
Competing interests
The authors declare that they have no competing interests.
Ethical approval
Ethical approval No. IR.SBMU.RICH.REC.1399.061 was granted by the Pediatric Infections Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Funding
The research reported in this publication was supported by the Researcher Grant Committee under grant number [20326] from the Pediatric Infections Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Authors’ ORCID
- Azimi L: https://orcid.org/0000-0002-7216-2530
- Shirkavand F: https://orcid.org/0009-0001-5207-4883
- Armin S: https://orcid.org/0000-0002-4993-482X
- Karbasian F: https://orcid.org/0000-0003-2494-6231
- Khodaei H: https://orcid.org/0000-0002-2297-4733
References
[1] Pouladfar G, Sanaei Dashti A, Kadivar MR, Jafari M, Pourabbas B, Jamalidoust M. Evaluation of multiplex real-time PCR and WHO criteria for diagnosing childhood bacterial meningitis in a tertiary referral hospital in Iran. Arch Pediatr Infect Dis. 2022;10(3):e101822. DOI: 10.5812/pedinfect.101822[2] Tsang RSW. A Narrative Review of the Molecular Epidemiology and Laboratory Surveillance of Vaccine Preventable Bacterial Meningitis Agents: Streptococcus pneumoniae, Neisseria meningitidis, Haemophilus influenzae and Streptococcus agalactiae. Microorganisms. 2021 Feb 22;9(2):449. DOI: 10.3390/microorganisms9020449
[3] Ranjbar R, Afshar D. Isothermal and sensitive identification of Streptococcus pneumoniae using loop mediated isothermal amplification assay. Arch Pediatr Infect Dis. 2018;6(1):e61604. DOI: 10.5812/pedinfect.61604
[4] Oordt-Speets AM, Bolijn R, van Hoorn RC, Bhavsar A, Kyaw MH. Global etiology of bacterial meningitis: A systematic review and meta-analysis. PLoS One. 2018;13(6):e0198772. DOI: 10.1371/journal.pone.0198772
[5] Corless CE, Guiver M, Borrow R, Edwards-Jones V, Fox AJ, Kaczmarski EB. Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J Clin Microbiol. 2001 Apr;39(4):1553-8. DOI: 10.1128/JCM.39.4.1553-1558.2001
[6] Bahr NC, Boulware DR. Methods of rapid diagnosis for the etiology of meningitis in adults. Biomark Med. 2014;8(9):1085-103. DOI: 10.2217/bmm.14.67
[7] Karimi A, Rafiei Tabatabaei S, Azimi L, Almasian Tehrani N, Fallah F, Faghihian I. Tracing the Negative Results of Multiplex Real-Time PCR Assay for Diagnosis of Bacterial Pediatrics Meningitis. Can J Infect Dis Med Microbiol. 2023;2023:3502666. DOI: 10.1155/2023/3502666
[8] WHO. Health topics – Meningitis. Geneva: WHO. Available from: https://www.who.int/health-topics/meningitis#tab=tab_1
[9] Hancerli Torun S, Calıskan B, Somer A, Salman N, Guler N. A case report; purulent meningitis due to serotype 2 of Streptococcus pneumoniae. Arch Pediatr Infect Dis. 2013; 1(3):144-6. DOI: 10.5812/pedinfect.5314.
[10] Haddad-Boubaker S, Lakhal M, Fathallah C, Bouafsoun A, Kharrat M, Khemiri M, Kechrid A, Smaoui H. Molecular diagnosis of bacterial meningitis by multiplex real time PCR in Tunisian children. J Infect Dev Ctries. 2018 Apr;12(4):235-43. DOI: 10.3855/jidc.9650
[11] Sacace Biotechnologies. NHS Meningitidis Real-TM Handbook. Real time PCR kit for detection of Neisseria meningitidis, Haemophilus influenzae and Streptococcus pneumoniae. Como, Italy: Sacace Biotechnologies; 2013 Mar 21. p. 16. Available from: http://bio-lab.com.pl/wp-content/uploads/2014/06/b2a78_NHS_Meningitis_Real_TM_21032013.pdf




